How to compare two germplasm and how to find varients from the interested region
Overview
Teaching: 10 min
Exercises: 10 minQuestions
How to search for sequence varients between two selected germplasm?
How to view genome within a given region?
Objectives
Providing a step by step demonstration on how to compare two germplasm and how to find sequence varients in an interested region.
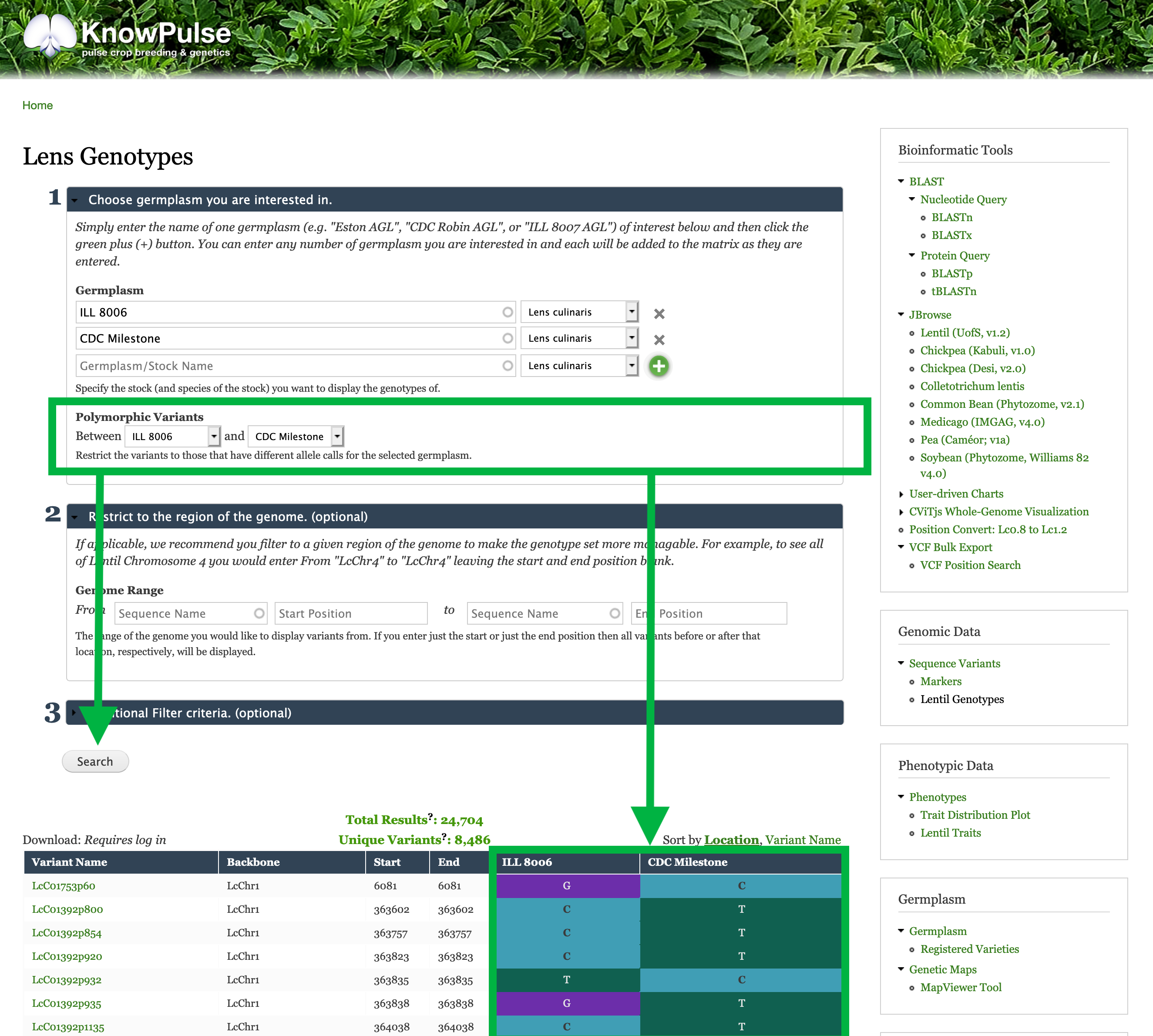
Restrict the Sequence Variants to polymorphic between your germplasm
Underneath germplasm, there is a filter to restrict to polymorphic variants. This filter compares two germplasm and only shows variants with different genotypic calls.
For our example, we would select ILL 8006 in the first drop down and CDC Milestone in the second drop-down to see only sequence variants with differing genotypes (i.e polymorphic variants) between these two germplasm.
Click Search to see the results.
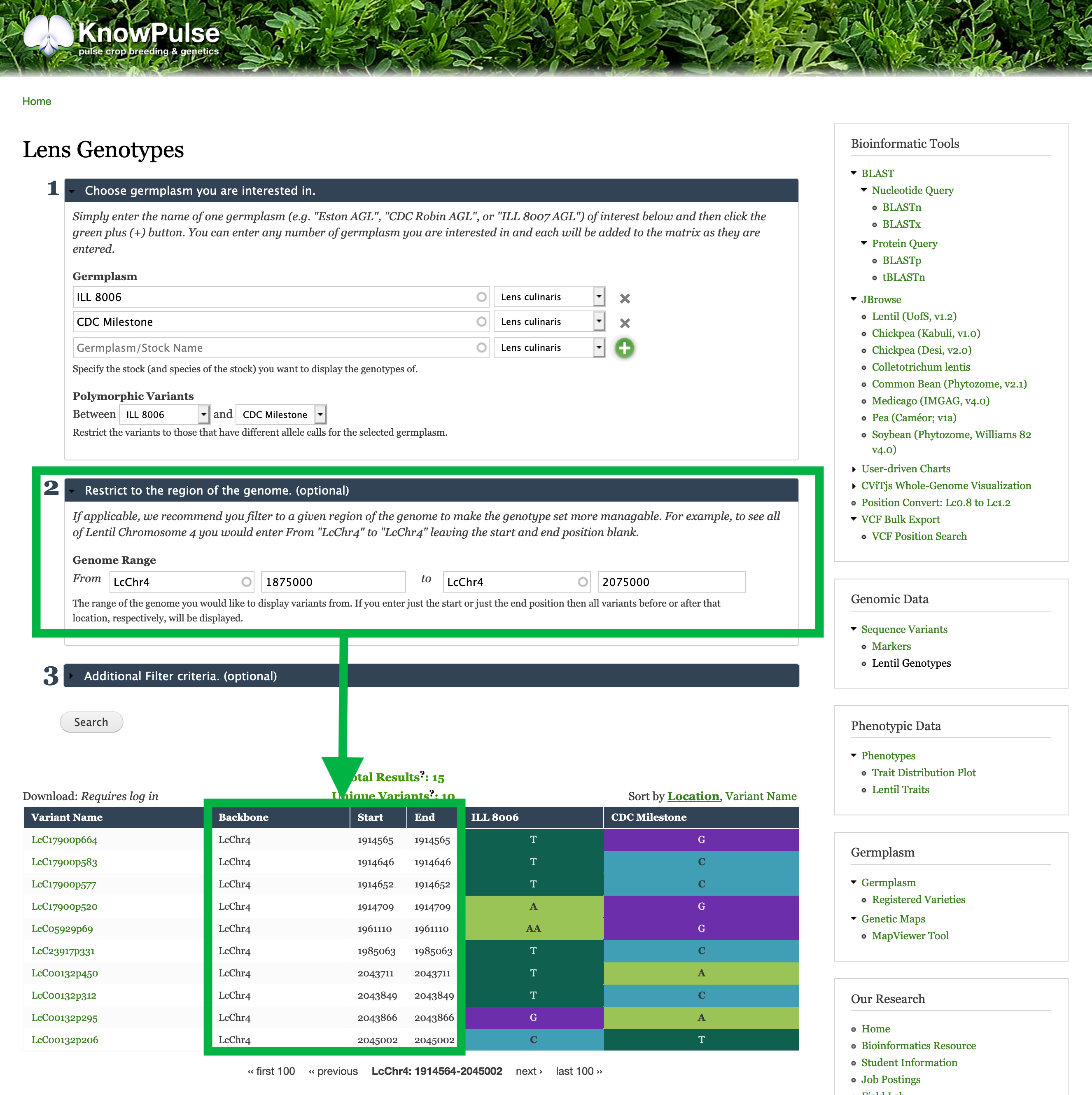
Restrict to your trait-implicated Region of the Genome
The second section of the filter criteria available for the genotype matrix allows you to enter the region of the genome you are interested in. Once you click search, the genotype matrix will only show sequence variants found in this region.
In our example, the region of interest is LcChr4:1875000-2075000. To enter this, we place LcChr4 for the Sequence Name, 1875000 for the start position and 2075000 for the end position.
Key Points
Log in KnowPulse with your user account before searching.